FLARE Users Guide and Tutorial
4/23/2020
Background
Requirements
1: Set up
2: Configure simulation (GLM)
3: Run your first simulation (GLM)
4: Examining FLARE output
5: Running GLM-AED
6: Modifying FLARE
7: Modifying FLARE for a new lake
Appendix: FLARE Configurations
Background
This document serves as a users guide and a tutorial for the FLARE (Forecasting Lake and Reservior Ecosystems) system (Thomas et al. 2020). FLARE generates forecasts and forecast uncertainty of water temperature and water quality for a 16-day time horizon at multiple depths of a lake or reservior. It uses data assimilation to update the initial starting point for a forecast and the model parameters based a real-time statistical comparsions to observations. While it has been developed, tested, and evaluated for Falling Creek Reservior in Vinton,VA (Thomas et al. 2020), this document provides guidance for adapting FLARE to different waterbodies.
FLARE is a set of R scripts that
- Downloads, cleans, organizes data required for data assimilation and forecasting
- Generating the inputs and configuration files required by the General Lake Model (GLM)
- Applying data assimilation to GLM
- Processing and archiving forecast output
- Visualizing forecast ouptut
FLARE uses the 1-D General Lake Model (Hipsey et al. 2019) as the mechanistic process model that predicts hydrodynamics of the lake or reservior. For forecasts of water quality, it uses GLM with the Aquatic Ecosystem Dynamics library. The binaries for GLM and GLM-AED are included in the FLARE code that is avialable on GitHub. FLARE requires GLM version 3.1 or higher.
More information about the GLM can be found here:
FLARE development has been supported by grants from National Science Foundation (CNS-1737424, DEB-1753639, EF-1702506, DBI-1933016, DEB-1926050)
Requirements
- RStudio
- R packages
- Tidyverse
- uses tidyr version 1.0.0 or higher (uses
pivot_longerandpivot_wider)
- uses tidyr version 1.0.0 or higher (uses
- mvtnorm
- ncdf4
- testit
- imputeTS
- tools
- rMR
- Tidyverse
1: Set up
To set up the files needed for FLARE perform either Step 1a or Step 1b
- Step 1a is the quickest way to get FLARE set-up but won’t have the most recent data or code.
- Step 1b is required if you want to automatically pull new data from GitHub
- Step 1c is uses a docker container that already has the libraries that FLARE requires installed. Similar to step 1a, the Docker container won’t have the most recent data or code.
1a: Pre-packaged code and data
Download and unzip the file here:
https://www.dropbox.com/s/enl9srn5rx7ecxp/flare_users_guide_files.zip?dl=0
1b: Generating directories, downloading code, and downloading data manually
Download FLARE code
-
Open R Studio
-
Locate the terminal tab
-
In the terminal, create the location on your computer where you want to save files for the tutorial using the command
mkdir. For example:mkdir /Users/quinn/Dropbox/Research/SSC_forecasting/flare_trainingThis will be called your tutorial directory below
-
In the terminal, move to the new directory using the command
cd. For example:cd /Users/quinn/Dropbox/Research/SSC_forecasting/flare_training -
In the terminal tab run the following command to download the FLARE code from Github
git clone https://github.com/CareyLabVT/FLARE.gitAfter the cloning process finishes, you should see a new folder named
FLAREin your working directory
Download data
-
Create a new directory for data used by FLARE in the tutorial directory. First, change your current directory to the tutorial directory using
cdfrom the terminal tab (if you just ran the step above, you should already be in this directory). For example:cd /Users/quinn/Dropbox/Research/SSC_forecasting/flare_training -
Then, run the following line to create the directory:
mkdir SCC_data -
Change your directory to this new data directory using
cdcd SCC_data -
Download the met station data by running the following command in the terminal (this may take a while)
git clone -b carina-data --single-branch https://github.com/CareyLabVT/SCCData.git carina-data -
Download the NOAA forecasts for FCR by running the following command in the terminal
git clone -b fcre --single-branch https://github.com/CareyLabVT/noaa_gefs_forecasts.git fcre -
Download the catwalk data by running the following command in the terminal
git clone -b mia-data --single-branch https://github.com/CareyLabVT/SCCData.git mia-data -
Download the weir data by running the following command in the terminal
git clone -b diana-data --single-branch https://github.com/CareyLabVT/SCCData.git diana-data -
Download noaa forecasts by running the following command in the terminal
git clone -b fcre --single-branch https://github.com/CareyLabVT/noaa_gefs_forecasts.git fcre -
Download non-sensor data by running the following command in the terminal
git clone -b manual-data --single-branch https://github.com/CareyLabVT/SCCData.git manual-dataThe non-sensor data include
- CTD data from EDI
- SSS files
- Nutrient chemistry from EDI
- Weir data from EDI
-
Download the met station from EDI (this file is too big for Github so you have to get it directory from EDI)
- If you would like to do this manually, the file can be found at:
https://portal.edirepository.org/nis/mapbrowse?packageid=edi.389.4.
Download the file
Met_final_2015_2019.csvand move it to yourSCC_data/manual-datadirectory
- If you would like to do this manually, the file can be found at:
https://portal.edirepository.org/nis/mapbrowse?packageid=edi.389.4.
Download the file
Create directory for running FLARE
-
Create a directory in your working directory (
flare_training, not theSCC_datadirectory) usingmkdir. For example:mkdir /Users/quinn/Dropbox/Research/SSC_forecasting/flare_training/flare_simulation
Copy the following files from FLARE/example_configuration_files to
flare_simulation
initiate_forecast_example.Rglm3_woAED.nmlconfigure_FLARE.R
1c: Using a Docker Container
Below are the steps for running FLARE as a Docker Container. If you already have Docker Desktop on your computer you can skip the first step.
- Download and install Docker Desktop from Docker.com
- Open a Terminal window in Mac or Command Prompt window in Windows
- Run this command in Terminal or Command Prompt (this will take a
while to download)
docker run --name flare-dev -e PASSWORD=password -p 8787:8787 flareforecast/flare_scc_tutorial2 - Open a webbrower and enter the following address: http://localhost:8787
- Enter the following user name and password
- user: rstudio
- password: password
2: Configure simulation (GLM)
A FLARE simulation with AED requires three files that should be in your
flare_simulation directory (coppied to this location in the step
above). The configuration funcations are spread across the files. These
files are described in more detail below
initiate_forecast_example.Rglm3_woAED.nmlconfigure_FLARE.R
initiate_forecast_example.R
This file is the main script that launches the FLARE code and plots the output. In this tutorial you will modify the following variables (for now only modify the first three as specified)
data_location: This is theSCC_datadirectory (full path; you will need to change this to reflect the location of your directory)code_folder: This is theFLAREdirectory (full path; you will need to change this to reflect the location of your directory)forecast_location: This is theflare_simulationdirectory (full path; you will need to change this to reflect the location of your directory)execute_location: This is the same asforecast_locationunless you are executing the simulation in a different directory. I use this to execute the code on a RAM disk to save read-writes to my hard disk. You don’t need to do this in the tutorialrestart_file: This is the full path to the file that you want to use as initial conditions for the simulation. You will set this toNAif the simulation is not a continuation of a previous simulation.sim_name: a string with the name of your simulation. This will appear in your output file namesforecast_days: This is your forecast horizon. The max is16days. Set to0if only doing data assimilation with observed drivers.spin_up_days: set to zero. Don’t worry about this one.start_day_local: The date that your simulation starts. Uses the YYYY-MM-DD format (e.g., “2019-09-20”).2018-07-12is the first day in the SCC project (when the catwalk temperature data comes online)start_time_local: The time of day you want to start a forecast. Because GLM is a daily timestep model, the simulation will start at this time. It usesmm:hh:ssformat and must only be a whole hour. It is in the local time zone of the lake in standard time. It can be any hour if only doing data assimilation with observed drivers (forecast_days = 0). If forecasting (forecast_days > 0) it is required to match up with the availability of a NOAA forecast. NOAA forecasts are available at the following times GMT so you must select a local time that matches one of these times (i.e., 07:00:00 at FCR is the 12:00:00 GMT NOAA forecast).- 00:00:00 GMT
- 06:00:00 GMT
- 12:00:00 GMT
- 18:00:00 GMT
forecast_start_day_local: The date that you want forecasting to start in your simulation. Uses the YYYY-MM-DD format (e.g., “2019-09-20”). The difference betweenstart_time_localandforecast_start_day_localdetermines how many days of data assimilation occur using observed drivers before handing off to forecasted drivers and not assimilating dataforecast_sss_on: Only used in AED simulations for setting the SSS (hypolimnetic oxygenation system) to on in the forecast
initiate_forecast_example.R loads Rscripts and runs the FLARE code
with the EnKF function run_flare and the plotting code
plot_forecast.
glm3_woAED.nml
glm3_woAED.nml is the configuration file that is required by GLM. It
can be configurated to run only GLM or GLM + AED. This version is
already configured to run only GLM for FCR and you do not need to modify
it for the example simulation.
configure_FLARE.R
configure_FLARE.R has the bulk of the configurations for FLARE that
you will set once and reuse (unlike initiate_forecast_example.R which
changes when you want to forecast a new time period). The end of this
document describes all of the configurations in configure_FLARE.R.
Later in the tutorial, you will modify key configurations in
configure_FLARE.R
If you set up your directories and modified the
initiate_forecast_example.R as described above, then you will not need
to modify configure_FLARE.R to do a test simulation.
3: Run your first simulation (GLM)
To run your first FLARE simulation confirm that the following variables
are set in your initiate_forecast_example.R file:
restart_file <- NAsim_name <- "test"forecast_days <- 16start_day_local <- "2018-07-12"start_time_local <- "07:00:00"forecast_start_day_local <- "2018-07-15"
Now source initiate_forecast_example.R.
You will find that a working_directory directory is created in your
flare_simulation directory. This is were all the files for the
simulation are stored. After the simulation is finished all the
important files are moved to the flare_simulation directory so it is
fine to delete the working_directory.
Your console will show a lot of messages from R that are mostly associated with reading and writing files. You will know that you simulation has started to work when you see the timestep being printed out to the console. This step will take a while.
Once the simulation is complete you will find a PDF and a netcdf (.nc)
file in flare_simulation directory. The PDF is the plotted output and
the netcdf file is the FLARE output.
4: Examining FLARE output
First, you can view the PDF that is output automatically. The PDF includes the mean and 95% CI for all depths simulated. The observations are added if they are available. If parameters are fit in the EnKF, they are also shown.
Example of FLARE output
Second, the netcdf output FLARE can be analyzed. Netcdf files are binary so you can not open them without special software. R has useful functions for working with netcdf files. For example, this is how you would analyze the mean temperature output from FLARE.
nc_file <- "example_output/test_H_2018_07_12_2018_07_15_F_16_4302020_15_13.nc"
nc <- nc_open(nc_file)
The data is stored as follows
print(nc)
## File example_output/test_H_2018_07_12_2018_07_15_F_16_4302020_15_13.nc (NC_FORMAT_NETCDF4):
##
## 21 variables (excluding dimension variables):
## float temp[time,ens,z] (Contiguous storage)
## units: deg_C
## _FillValue: 1.00000003318135e+32
## long_name: temperature
## float temp_mean[time,z] (Contiguous storage)
## units: deg_C
## _FillValue: 1.00000003318135e+32
## long_name: temperature_mean
## float temp_upperCI[time,z] (Contiguous storage)
## units: deg_C
## _FillValue: 1.00000003318135e+32
## long_name: temperature_upperCI
## float temp_lowerCI[time,z] (Contiguous storage)
## units: deg_C
## _FillValue: 1.00000003318135e+32
## long_name: temperature_lowerCI
## float qt_restart[states_aug,states_aug] (Contiguous storage)
## units: -
## _FillValue: 1.00000003318135e+32
## long_name: restart covariance matrix
## float x_restart[ens,states_aug] (Contiguous storage)
## units: -
## _FillValue: 1.00000003318135e+32
## long_name: matrix for restarting EnKF
## float x_prior[time,ens,states_aug] (Contiguous storage)
## units: -
## _FillValue: 1.00000003318135e+32
## long_name: Predicted states prior to Kalman correction
## float obs[time,obs_dim] (Contiguous storage)
## units: various
## _FillValue: 1.00000003318135e+32
## long_name: temperature observations
## int forecasted[time] (Contiguous storage)
## units: -
## _FillValue: -99
## long_name: 0 = historical; 1 = forecasted
## float surface_height_restart[ens] (Contiguous storage)
## units: m
## _FillValue: -99
## long_name: Surface Height
## float snow_ice_restart[ens,snow_ice_dim] (Contiguous storage)
## units: m
## _FillValue: -99
## long_name: Snow (1), White Ice (2), Blue Ice (3)
## float ice_thickness[time,ens] (Contiguous storage)
## units: m
## _FillValue: -99
## long_name: Ice Thickness
## float lake_depth[time,ens] (Contiguous storage)
## units: m
## _FillValue: -99
## long_name: Depth of lake
## float avg_surf_temp_restart[ens] (Contiguous storage)
## units: deg_C
## _FillValue: -99
## long_name: Running Average of Surface Temperature
## float running_residuals[qt_update_days,states] (Contiguous storage)
## units: various
## _FillValue: 1.00000003318135e+32
## long_name: running residual for updating qt
## float mixing_restart[ens,mixing_restart_vars_dim] (Contiguous storage)
## units: various
## _FillValue: 1.00000003318135e+32
## long_name: variables required to restart mixing
## float depths_restart[ens,depth_restart_vars_dim] (Contiguous storage)
## units: various
## _FillValue: 1.00000003318135e+32
## long_name: depths simulated by glm that are required to restart
## float zone1temp[time,ens] (Contiguous storage)
## units: deg_C
## _FillValue: 1.00000003318135e+32
## float zone2temp[time,ens] (Contiguous storage)
## units: deg_C
## _FillValue: 1.00000003318135e+32
## float sw_factor[time,ens] (Contiguous storage)
## units: -
## _FillValue: 1.00000003318135e+32
## float inflow_factor[time,ens] (Contiguous storage)
## units: -
## _FillValue: 1.00000003318135e+32
##
## 10 dimensions:
## time Size:20
## units: seconds
## long_name: seconds since 1970-01-01 00:00.00 UTC
## ens Size:210
## long_name: ensemble member
## z Size:28
## units: meters
## long_name: Depth from surface
## states_aug Size:32
## long_name: length of model states plus parameters
## obs_dim Size:28
## long_name: length of
## snow_ice_dim Size:3
## long_name: snow ice dims
## qt_update_days Size:30
## long_name: Number of running days that qt smooths over
## states Size:28
## long_name: states
## mixing_restart_vars_dim Size:17
## long_name: number of mixing restart variables
## depth_restart_vars_dim Size:500
## long_name: number of possible depths that are simulated in GLM
##
## 6 global attributes:
## title: Falling Creek Reservoir forecast
## institution: Virginia Tech
## GLM_version: GLM 3.0.0beta10
## FLARE_version: v1.0_beta.1
## time_zone_of_simulation: EST
## forecast_issue_time: 2020-04-30 15:13:55
The key to understanding the file is the dimensions. For the variable
temp_mean the first dimension is time and the second is z, which
is depth. The actual values for these dimensions are in the variables
with in the dimensions called time and z. The other key dimension is
ens, which is the ensemble member. It is not present in temp_mean
because that output variable is summarized across ensemble members.
Plotting FLARE output
To visualize the output, you can use the following code
temp_mean <- ncvar_get(nc, varid = "temp_mean") #get the mean temperatures from the netcdf file
t <- ncvar_get(nc, varid = "time") #get the corresponding times from the netcdf file
t <- as.POSIXct(t, origin = '1970-01-01 00:00.00 UTC', tz = "EST") #format times as datetime
depths <- ncvar_get(nc, varid = "z")
focal_depth <- 1 #What depth are you interested in (meters)
depth_index <- which.min(abs(depths - 1))
d <- tibble(time = t, #create a dataframe of times and temperatures at the specified depth
temp_mean = temp_mean[,depth_index])
ggplot(d, aes(x = time, y = temp_mean)) + #plot mean temperature (modeled) at this depth over time
geom_line() +
geom_point() +
labs(x = "Date", y = expression(~degree~C), title = "FCR forecast")
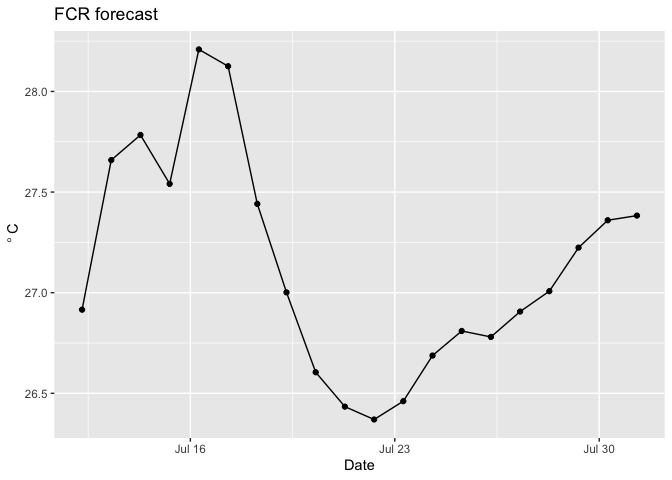
However, you do not know which points were based on observed drivers and
data assimilation and which points were forecasted. To find the points
that are forecasted you need the forecasted variable in the
output.
forecasted <- ncvar_get(nc, varid = "forecasted") #binary. 1: forecasted, 0: not forecasted
focal_depth <- 1 #What depth are you interested in (meters)
depth_index <- which.min(abs(depths - 1))
d <- tibble(time = t,
temp_mean = temp_mean[,depth_index],
forecasted = as.logical(forecasted))
ggplot(d) +
geom_line(aes(x = time, y = temp_mean)) +
geom_point(aes(x = time, y = temp_mean, color = forecasted)) +
labs(x = "Date", y = expression(~degree~C), title = "FCR forecast")
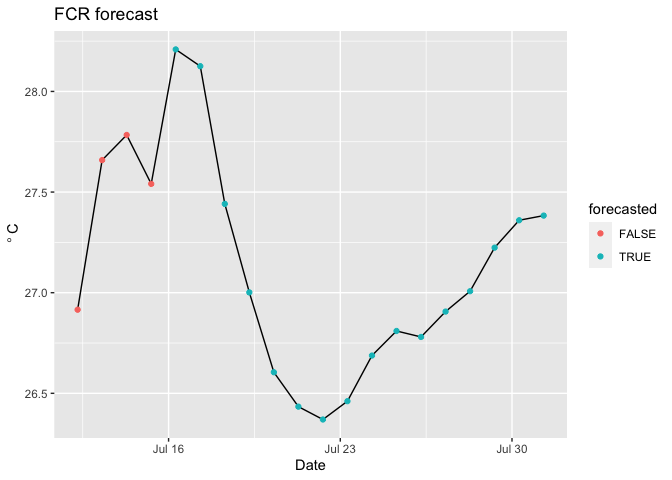
You can add the uncertainty using the following.
temp_mean <- ncvar_get(nc, varid = "temp_mean")
temp_upper95 <- ncvar_get(nc, varid = "temp_upperCI")
temp_lower95 <- ncvar_get(nc, varid = "temp_lowerCI")
forecasted <- ncvar_get(nc, varid = "forecasted")
t <- ncvar_get(nc, varid = "time")
depths <- ncvar_get(nc, varid = "z")
t <- as.POSIXct(t, origin = '1970-01-01 00:00.00 UTC', tz = "EST")
focal_depth <- 1 #What depth are you interested in (meters)
depth_index <- which.min(abs(depths - 1))
d <- tibble(time = t,
temp_mean = temp_mean[,depth_index],
temp_upper95 = temp_upper95[,depth_index],
temp_lower95 = temp_lower95[,depth_index],
forecasted = as.logical(forecasted))
ggplot(d,aes(x = time)) +
geom_line(aes(y = temp_mean)) +
geom_ribbon(aes(ymin = temp_lower95, ymax = temp_upper95), alpha = 0.25) +
geom_vline(xintercept = last(d$time[which(d$forecasted == 0)])) + #vertical line marking the day of the forecast
labs(x = "Date", y = expression(~degree~C), title = "FCR forecast")
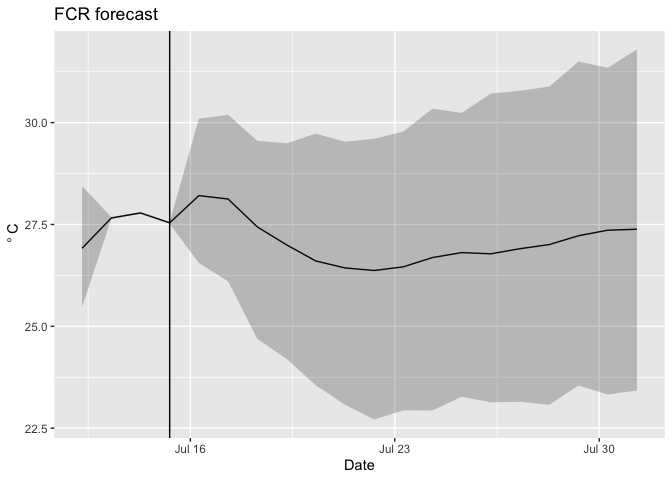
You can add observations
temp_mean <- ncvar_get(nc, varid = "temp_mean")
temp_upper95 <- ncvar_get(nc, varid = "temp_upperCI")
temp_lower95 <- ncvar_get(nc, varid = "temp_lowerCI")
forecasted <- ncvar_get(nc, varid = "forecasted")
t <- ncvar_get(nc, varid = "time")
depths <- ncvar_get(nc, varid = "z")
t <- as.POSIXct(t, origin = '1970-01-01 00:00.00 UTC', tz = "EST")
obs <- ncvar_get(nc, varid = "obs")
focal_depth <- 1 #What depth are you interested in (meters)
depth_index <- which.min(abs(depths - 1))
d <- tibble(time = t,
temp_mean = temp_mean[,depth_index],
temp_upper95 = temp_upper95[,depth_index],
temp_lower95 = temp_lower95[,depth_index],
forecasted = as.logical(forecasted),
obs = obs[, depth_index])
ggplot(d,aes(x = time)) +
geom_line(aes(y = temp_mean)) +
geom_ribbon(aes(ymin = temp_lower95, ymax = temp_upper95), alpha = 0.25) +
geom_point(aes(y = obs), color = "red") +
geom_vline(xintercept = last(d$time[which(d$forecasted == 0)])) +
labs(x = "Date", y = expression(~degree~C), title = "FCR forecast")
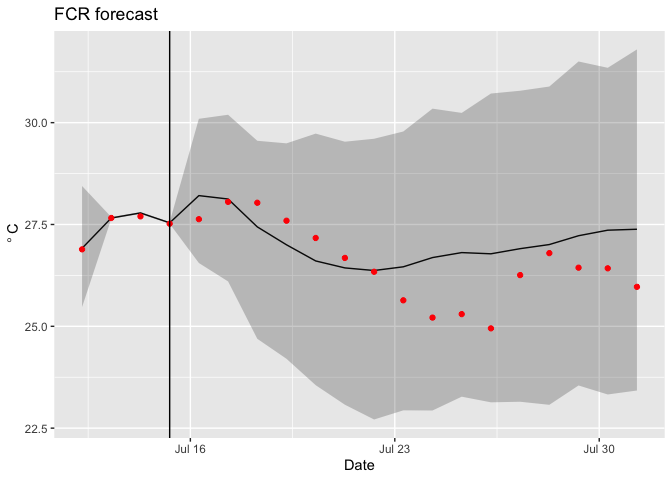
Finally, you can calculate the statistics of the forecast directly from the ensembles (rather than using the statistics already calculated in the netcdf file; these should be identical for mean and 95% confidence intervals)
temp <- ncvar_get(nc, varid = "temp")
forecasted <- ncvar_get(nc, varid = "forecasted")
t <- ncvar_get(nc, varid = "time")
t <- as.POSIXct(t, origin = '1970-01-01 00:00.00 UTC', tz = "EST")
depths <- ncvar_get(nc, varid = "z")
obs <- ncvar_get(nc, varid = "obs")
focal_depth <- 1 #What depth are you interested in (meters)
depth_index <- which.min(abs(depths - 1))
temp_lower95 <- rep(NA, length(t))
temp_upper95 <- rep(NA, length(t))
temp_mean <- rep(NA, length(t))
for(i in 1:length(t)){
temp_lower95[i] <- quantile(temp[i, , depth_index],0.025) #calculate the 95% confidence interval from all ensemble members at each date
temp_upper95[i] <- quantile(temp[i, , depth_index],0.975)
temp_mean[i] <- mean(temp[i, , depth_index]) #calculate the mean of all ensemble members at each date
}
d <- tibble(time = t,
temp_mean = temp_mean,
temp_upper95 = temp_upper95,
temp_lower95 = temp_lower95,
forecasted = as.logical(forecasted),
obs = obs[, depth_index])
d
## # A tibble: 20 x 6
## time temp_mean temp_upper95 temp_lower95 forecasted obs
## <dttm> <dbl> <dbl> <dbl> <lgl> <dbl>
## 1 2018-07-12 07:00:00 26.9 28.4 25.5 FALSE 26.9
## 2 2018-07-13 07:00:00 27.7 27.7 27.6 FALSE 27.7
## 3 2018-07-14 07:00:00 27.8 27.8 27.8 FALSE 27.7
## 4 2018-07-15 07:00:00 27.5 27.6 27.5 FALSE 27.5
## 5 2018-07-16 07:00:00 28.2 30.1 26.6 TRUE 27.6
## 6 2018-07-17 07:00:00 28.1 30.2 26.1 TRUE 28.1
## 7 2018-07-18 07:00:00 27.4 29.6 24.7 TRUE 28.0
## 8 2018-07-19 07:00:00 27.0 29.5 24.2 TRUE 27.6
## 9 2018-07-20 07:00:00 26.6 29.7 23.6 TRUE 27.2
## 10 2018-07-21 07:00:00 26.4 29.5 23.1 TRUE 26.7
## 11 2018-07-22 07:00:00 26.4 29.6 22.7 TRUE 26.3
## 12 2018-07-23 07:00:00 26.5 29.8 22.9 TRUE 25.6
## 13 2018-07-24 07:00:00 26.7 30.3 22.9 TRUE 25.2
## 14 2018-07-25 07:00:00 26.8 30.2 23.3 TRUE 25.3
## 15 2018-07-26 07:00:00 26.8 30.7 23.1 TRUE 24.9
## 16 2018-07-27 07:00:00 26.9 30.8 23.1 TRUE 26.3
## 17 2018-07-28 07:00:00 27.0 30.9 23.1 TRUE 26.8
## 18 2018-07-29 07:00:00 27.2 31.5 23.5 TRUE 26.4
## 19 2018-07-30 07:00:00 27.4 31.3 23.3 TRUE 26.4
## 20 2018-07-31 07:00:00 27.4 31.8 23.4 TRUE 26.0
5: Running GLM-AED
To run GLM-AED fist copy a glm namelist file to your simulation
flare_simulation directory. Thre are two example options in the
FLARE/example_configuration_files directory. They differ in whether
they include an additional inflow and outflow that represents the SSS
(bottom-water oxygenation system).
glm3_wAED_wSSS.nml: Include the SSS inflows and outflowsglm3_wAED_woSSS.nml: Excludes the SSS inflows and outflows
Second, copy an aed namelist file to your simulation flare_simulation
directory. Thre are two example options in the
FLARE/example_configuration_files directory. They differ in whether
they include all the water quality variables in AED or only oxygen.
aed2.nml: All variablesaed2_only_Oxy.nml: Only oxygen
Third, modify the following in configure_FLARE.R:
include_wq: Set toTRUEbase_GLM_nml: full path to the glm namelist matches the one in yourflare_simulationthat includes AED.base_AED_nml: full path to your aed namelistbase_AED_phyto_pars_nml: full path to your phyto_pars namelistbase_AED_zoop_pars_nml: full path to your zoop_pars namelist
Finally, confirm that the following varibles in configure_FALRE.R
match your desire use of the data:
do_methods: these are the names of the different measurement methods as defined in the temp_oxy_chla_qaqc.R and extract_CTD.R scripts- current values for FCR:
exo,do,ctd
- current values for FCR:
chla_methods: these are the names of the different measurement methods as defined in the temp_oxy_chla_qaqc.R and extract_CTD.R scripts- current values for FCR:
exo,ctd
- current values for FCR:
fdom_methods: these are the names of the different measurement methods as defined in the temp_oxy_chla_qaqc.R and extract_nutrients.R scripts- current values for FCR:
exo,grap_sample
- current values for FCR:
nh4_methods: these are the names of the different measurement methods as defined in the extract_nutrients.R- current values for FCR:
grap_sample
- current values for FCR:
no3_methods: these are the names of the different measurement methods as defined in the extract_nutrients.R- current values for FCR:
grap_sample
- current values for FCR:
srp_methods: these are the names of the different measurement methods as defined in the extract_nutrients.Rtime_threshold_seconds_oxygen: this is the number of seconds that an observation has to be within thestart_time_localto be used in the analysistime_threshold_seconds_chla: this is the number of seconds that an observation has to be within thestart_time_localto be used in the analysistime_threshold_seconds_fdom: this is the number of seconds that an observation has to be within thestart_time_localto be used in the analysistime_threshold_seconds_nh4: this is the number of seconds that an observation has to be within thestart_time_localto be used in the analysistime_threshold_seconds_no3: this is the number of seconds that an observation has to be within thestart_time_localto be used in the analysistime_threshold_seconds_srp: this is the number of seconds that an observation has to be within thestart_time_localto be used in the analysis
6: Modifying FLARE
Turning off data assimilation
- Our for modification to FLARE will be remove a source of uncertainty
in the forecast The
uncert_modeallows you to do this easily based on the following:- 1 = all types
- 2 = no uncertainty
- 3 = only process uncertainty
- 4 = only NOAA weather forecast uncertainty
- 5 = only initial condition uncertainty
- 6 = only initial condition uncertainty and no state updating with EnKF
- 7 = only parameter uncertainty
- 8 = only meteorology downscaling uncertainty
- 9 = no sources of uncertainty and no state updating with EnKF
- 11 = all sources of uncertainty and no state updating with EnKF
In this tutorial, explore how data assimilation influences your forecast. To do this modify the following two variables and source the code:
uncert_mode <- 11(inconfigure_FLARE.R)sim_name <- "NO_DA"(ininitiate_forecast_example.R)- source
initiate_forecast_example.R
Removing parameter estimation
You can remove the parameter optimization so that FLARE only uses data
assimilation to update the initial conditions of a forecast. * In
configure_FLARE.R add the following code to line 250. These remove the
parameters from EnKF.
par_names <<- c()
par_names_save <<- c()
par_nml <<- c()
par_init_mean <<- c()
par_init_lowerbound <<- c()
par_init_upperbound <<- c()
par_lowerbound <<- c()
par_upperbound <<- c()
par_init_qt <<- c()
par_units <<- c()
Increasing observational uncertainty
The second modification you will do is to to increase the observational
uncertainty. In configure_FLARE.R set obs_error_temperature_intercept
= 1. Then Source initiate_forecast_example.R. Importantly,
observational uncertainty is in variance units (the square of standard
deviation).
Observational uncertainty has two components.
obs_error_temperature_intercept: the component that is independent of the measurement magnitudeobs_error_temperature_slope: the component that linearly scales with the magnitude of the measurement
Changing the ensemble size
The variable ensemble_size allows you to adjust the size of the
ensemble. You can use any value if you are using data assimilation with
observed drivers. If you are forecasting then ensemble_size must be a
multiple of 21 * n_ds_members, where 21 is the number of NOAA GEF
ensemble members.
Changing the number of depths simulated
The variable modeled_depths allows you to adjust the depths that FLARE
simulates
7: Modifying FLARE for a new lake
We are in the process of making it easier to apply FLARE to a new lake. Currently applying FLARE to a new lake requires modifying R scripts because data streams (meteorology, inflow, and in situ data) formats differ between lakes. The following steps are required to set up a new lake:
Modify the glm namelist
Within the glm namelist (glm3_woAED.nml in the tutorial), at minimum
you need to change the variables in the &morphometry, &inflow, and
&outflow sections. Please see the GLM users
guide for
more information about the variables in these sections of the namelist.
Additionally, the paper describing
GLM 3.0.0 describes
variables in the namelist
You will also need to modify the zone_heights in the &sediment
section for your new lake. Currently, FLARE only handles two
zone_heights, where the first value is the top depth of the bottom
layer and the second value is the top of the top layer (the maximum
depth of the lake).
Other variables to modify are:
Kw: the light extension coefficient
The FLARE code automatically updates all the variables in
&init_profiles
Modify Configure_flare.R
Within configure_flare.R you will need to update:
lake_name: this is the code for the lake. It must match the the lake specific directory within your/FLARE/Rscripts/[lake_name]directory.lake_latitude: Degrees Northlake_longitude: Degrees Westlocal_tzone: Standard time, a time zone that R understandslake_depth_init: Initial depth of the lakemodeled_depths: depths that are simulated
You will also need to update the file names for the driver and in situ data:
insitu_obs_fname: the in situ data (temperature, do, etc)met_obs_fname: the meteorological stationinflow_file1: the first inflow (the weir at FCR)inflow_file2: an optional second inflowoutflow_file1: an optional outflow (NOTE: this currently is not used in FLARE)
FLARE also allows you to include a separate file for CTD depth profiles:
ctd_fname: a value of NA is needed if you do not have or do not want to use CTD measurements
Importantly, insitu_obs_fname, met_obs_fname, and inflow_file1
require two file names. The first is the realtime file that has not
had QAQC applied (i.e,just downloaded from the sensor). The second is
file that has QAQC applied (i.e, a file from a data repository). If any
time periods overlap between the two files, FLARE will default the
second (QAQCed) file. Historical data assimilation applications will use
the QAQCed file. If you are missing one of the two file, the vector
should have an NA in its place.
You will need to specify how to process data files:
temp_methods: these are the names of the different measurement methods used to aquire data (e.g. CTD profile, in situ thermistor), as defined in the temp_oxy_chla_qaqc.R and, potentially, extract_CTD.R, scripts that you have modified for the new lake.- current values for FCR:
thermistor,exo,do,ctd
- current values for FCR:
time_threshold_seconds_temp: this is how close a measurement needs to be to the forecast time (start_time_local) to be used in the analysis, in seconds.distance_threshold_meter: this is the distances in meters that an observation has to be within to be matched to a value inmodeled_depths.
Finally there are some plotting configurations:
focal_depths_manager: the depths that are included in the plot with the % chance of turnoverturnover_index_1: the top depth used in the calculation of whether the lake is mixedturnover_index_2: the bottom depth used in the calculation of whether the lake is mixed
Creating and modifying a lake-specific Rscripts directory
You need to create a /FLARE/Rscripts/[lake_name] directory and move
all the files in /FLARE/Rscripts/fcre to that directory.
You may need to modify the following files that you moved to
/FLARE/Rscripts/[lake_name], depending on how similar your data file
formats are to the files used at fcre.
met_qaqc.R:- Inputs:
fnamecleaned_met_fileinput_file_tzlocal_tzonefull_time_local
- Outputs: needs to write a csv file with the following columns
- timestamp
- ShortWave
- LongWave
- AirTemp
- RelHum
- WindSpeed
- Rain
- Inputs:
inflow_qaqc.R:- Inputs:
realtime_fileqaqc_filenutrients_filecleaned_inflow_filelocal_tzoneinput_file_tz
- Output: needs to write a csv file with the following columns
- GLM only: time, FLOW, TEMP, SALT
- GLM AED: variables defined in
wq_names(configure_FLARE.R)
- Inputs:
in_situ_qaqc.R: This function calls the following functions that need to be modified based on the format of the lake data streams- Inputs:
insitu_obs_fnamedata_locationmaintenance_filectd_fnamenutrients_fnamecleaned_observations_file_longlake_namecode_folder
- Outputs: needs to write a csv file with the following columns
- timestamp: date-time class
- depth: meters
- variable: measurement variable
- method: user defined string defining the type of
measurement. Matches methods in
configure_FLARE.R(i.e.,temp_methods)
in_situ_qaqc.Rcalls the following functionstemp_oxy_chla_qaqc.R:- Inputs:
realtime_fileqaqc_filemaintenance_fileinput_file_tz
- Output: data frame with the following columns
- timestamp: date-time class
- depth: meters
- variable: measurement variable
- method: user defined string defining the type of
measurement. Matches methods in
configure_FLARE.R(i.e.,temp_methods)
- Inputs:
extract_CTD.R:- Inputs:
fnameinput_file_tzlocal_tzone
- Output: data frame with the following columns
- timestamp: date-time class
- depth: meters
- variable: measurement variable
- method: user defined string defining the type of
measurement. Matches methods in
configure_FLARE.R(i.e.,temp_methods)
- Inputs:
extract_nutrients.R:- Inputs:
fnameinput_file_tzlocal_tzone
- Output: data frame with the following columns
- timestamp: date-time class
- depth: meters
- variable: measurement variable
- method: user defined string defining the type of
measurement. Matches methods in
configure_FLARE.R(i.e.,temp_methods)
- Inputs:
- Inputs:
Appendix: FLARE Configurations
A guide to the variables in configure_flare.R
General set-up
GLMversion: string of GLM versionFLAREversion: string of FLARE versionuse_null_model: Whether to use the persistance null model or GLMTRUE: use persistance null modelFALSE: use GLM
include_wq: Whether to use GLM-AED or just GLMTRUE: use GLM-AEDFALSE: only use GLM
uncert_mode: code for the type of uncertainty to include in simulation- 1 = all types of uncertainty
- 2 = no uncertainty
- 3 = only process uncertainty
- 4 = only NOAA weather forecast uncertainty
- 5 = only initial condition uncertainty
- 6 = only initial condition uncertainty and no state updating with the EnKF
- 7 = only parameter uncertainty
- 8 = only meteorology downscaling uncertainty
- 9 = no sources of uncertainty and no state updating with EnKF
- 11 = all sources of uncertainty and no state updating with EnKF
single_run: Removes uncertainty and only simulates 3 ensemble membersTRUE:: only simulates 3 ensemble members without uncertaintyFALSE: all ensemble members and uncertainty specified inuncert_mode
pull_from_git: Flag to pull data from GitHubTRUE: PullFALSE: Don’t pull (needed if you are offline)
push_to_git: Push results to GitHubTRUE: PushFALSE: Don’t Push (needed if you are offline)
Lake specific variables
lake_name: four letter code name for lake. Match lake directy inFLARE/Rscriptsand in the NOAA GEF fileslake_latitude: Degrees Northlake_longitude: Degrees Westlocal_tzone: In standard time. Must be recognized by R.
Weather forcing options
use_future_metTRUE: use NOAA forecast for “Future”FALSE= use observed weather for “Future”; only works if “forecasting” past dates
DOWNSCALE_MET: apply spatial and temporal downscalingTRUE: downscaleFALSE: don’t downscale
noaa_location: full path of directory with NOAA GEF files- paste0(data_location, “/”,lake_name,“/”)
downscaling_coeff: full path to the Rdata that has previously estimated downscaling coefficients- paste0(data_location, “/manual-data/debiased.coefficients.2018_07_12_2019_07_11.RData”)
- NA: no previous coefficient, calculate within simulation if
DOWNSCALE_MET == TRUE.
met_ds_obs_start: Starting date to include in estimation of downscaling coefficients- date class (
as.Date("2019-07-11"))
- date class (
met_ds_obs_end: Ending date to include in estimation of downscaling coefficients- date class (
as.Date("2019-07-11"))
- date class (
missing_met_data_threshold: PENDING
Inflow options
use_future_inflow: Use forecast inflow vs. observed inflow (if available)TRUE: Future inflowFALSE: Observed inflow
future_inflow_flow_coeff: Vector of three numbers- Intercept
- Coefficient with lagged flow rate
- Coefficient with lagged rain
future_inflow_flow_error: Standard deviation of future flow modelfuture_inflow_temp_coeff:- Intercept
- Coefficient with lagged water temperature
- Coefficient with lagged air temperature
future_inflow_temp_error: Standard deviation of future temperature model
GLM namelist files
base_GLM_nml: full path to the glm namelist- example: paste0(forecast_location,“/glm3_woAED.nml” )
base_AED_nml: full path to the aed namelistbase_AED_phyto_pars_nml: full path to the phyto_pars namelistbase_AED_zoop_pars_nml: full path to the zoop_pars namelist
Depth information
modeled_depths: Vector of depths are that represented in the EnKF
Ensemble description
ensemble_size: Total number of ensemble membersn_ds_members: Number of random samples drawn from each NOAA GEF ensemble member based on the variance-covariance matrix from downscaling analysis.n_inflow_outflow_members: Number of random samples drawn from the normal distribution describing the residual error in the inflow rate and inflow temperature model
Process uncertainty adaption
use_cov: Allow for covariance of process uncertainty across depthsTRUE: IncludeFALSE: Don’t include
adapt_qt_method:- 0 = no adapt,
- 1 = variance in residuals
num_adapt_days: Number of days included in generation of the variance- covariance matrix of the residuals. The matrix represents process uncertainties.Inflat_pars: The variance inflation factor applied to the parameter component of the ensemble. Value greater than 1.
Parameter calibration information
par_names: vector of GLM names of parameter values estimatedpar_names_save: vector of names of parameter values estimated that are desired in output and plotspar_nml: vector of nml file names that contains the parameter that is being estimatedpar_init_mean: vector of initial mean value for parameterspar_init_lowerbound: vector of lower bound for the initial uniform distribution of the parameterspar_init_upperbound: vector of upper bound for the initial uniform distribution of the parameterspar_lowerbound: vector of lower bounds that a parameter can havepar_upperbound: vector of upper bounds that a parameter can havepar_init_qt: Not usedpar_units: Units of parameter for plotting
Observation information
-
realtime_insitu_location: full path to the directory with the non-QAQCed in situ data (i.e., thermistor chain) -
realtime_met_station_location: full path to the directory with the non-QAQCed met station data -
manual_data_location: full path to the directory with the QAQCed datasets -
realtime_inflow_data_location: full path to the directory with the non-QAQCed inflow data. -
ctd_fname: full path of CTD file -
nutrients_fname: full path of nutrients file -
insitu_obs_fname: vector of two full paths- 1: path of the realtime file (required)
- 2: path of respository (QAQCed) file (if not used, must have
NA)
-
variable_obsevation_depths: Use the depth measurement from the exo sonde (on a buoy; constant depth from surface) to convert the depths of a fixed thermistor chain to distance from surfaceTRUE: convertFALSE: don’t convert
-
ctd_2_exo_chla: slope and intercept of linear regression that converts chla values from the CTD to values from the exo sonde -
met_obs_fname: vector of two full paths- 1: path of the realtime file (required)
- 2: path of respository (QAQCed) file (if not used, must have
NA)
-
inflow_file1: vector of two full paths- 1: path of the realtime file (required)
- 2: path of respository (QAQCed) file (if not used, must have
NA)
-
outflow_file1: Not current used -
inflow_file2: Not current used -
temp_methods: these are the names of the different measurement methods as defined in thetemp_oxy_chla_qaqc.Randextract_CTD.Rscripts- current values for FCR:
thermistor,exo,do,ctd
- current values for FCR:
-
do_methods: these are the names of the different measurement methods as defined in thetemp_oxy_chla_qaqc.Randextract_CTD.Rscripts- current values for FCR:
exo,do,ctd
- current values for FCR:
-
chla_methods: these are the names of the different measurement methods as defined in thetemp_oxy_chla_qaqc.Randextract_CTD.Rscripts- current values for FCR:
exo,ctd
- current values for FCR:
-
fdom_methods: these are the names of the different measurement methods as defined in thetemp_oxy_chla_qaqc.Randextract_nutrients.Rscripts- current values for FCR:
exo,grap_sample
- current values for FCR:
-
nh4_methods: these are the names of the different measurement methods as defined in theextract_nutrients.R- current values for FCR:
grap_sample
- current values for FCR:
-
no3_methods: these are the names of the different measurement methods as defined in theextract_nutrients.R- current values for FCR:
grap_sample
- current values for FCR:
-
srp_methods: these are the names of the different measurement methods as defined in theextract_nutrients.R -
time_threshold_seconds_temp: this is how close a measurement needs to be to the forecast time (start_time_local) to be used in the analysis, in seconds. -
time_threshold_seconds_oxygen: this is how close a measurement needs to be to the forecast time (start_time_local) to be used in the analysis, in seconds. -
time_threshold_seconds_chla: this is how close a measurement needs to be to the forecast time (start_time_local) to be used in the analysis, in seconds. -
time_threshold_seconds_fdom: this is how close a measurement needs to be to the forecast time (start_time_local) to be used in the analysis, in seconds. -
time_threshold_seconds_nh4: this is how close a measurement needs to be to the forecast time (start_time_local) to be used in the analysis, in seconds. -
time_threshold_seconds_no3: this is how close a measurement needs to be to the forecast time (start_time_local) to be used in the analysis, in seconds. -
time_threshold_seconds_srp: this is how close a measurement needs to be to the forecast time (start_time_local) to be used in the analysis, in seconds. -
distance_threshold_meter: this is the distances in meters that an observation has to be within to be matched to a value inmodeled_depths.
Initial Conditions (GLM)
lake_depth_init: initial lake depth (meters)default_temp_init: vector of initial temperature profiledefault_temp_init_depths: vector of depths in initial temperature profilethe_sals_init: vector of initial salinty valuesdefault_snow_thickness_init: initial snow thickness (cm)default_white_ice_thickness_init: initial white ice thickness (cm)default_blue_ice_thickness_init: initial blue ice thickness (cm)
Water quality variable information
tchla_components_vars: The names of the phytoplankton groups that contribute to total chl-a in GLMwq_names: GLM names of the water quality variables modeled. Not used ifinclude_wq == FALSEbiomass_to_chla: Carbon to chlorophyll ratio (mg C/mg chla) for each group intchla_components_vars.- 12 g/ mole of C vs. X g/ mole of chla
init_donc: Initial ratio of DON:DOCinit_dopc: Initial ratio of DOP:DOCinit_ponc: Initial ratio of PON:POCinit_popc: Initial ratio ofOXY_oxy_init: Initial concentration (mmol/m3)CAR_pH_init: Initial concentration (mmol/m3)CAR_dic_init: Initial concentration (mmol/m3)CAR_ch4_init: Initial concentration (mmol/m3)SIL_rsi_init: Initial concentration (mmol/m3)NIT_amm_init: Initial concentration (mmol/m3)NIT_nit_init: Initial concentration (mmol/m3)PHS_frp_init: Initial concentration (mmol/m3)OGM_doc_init: Initial concentration (mmol/m3)OGM_poc_init: Initial concentration (mmol/m3)OGM_pon_init: Initial concentration (mmol/m3)OGM_pop_init: Initial concentration (mmol/m3)NCS_ss1_init: Initial concentration (mmol/m3)PHS_frp_ads_init: Initial concentration (mmol/m3)PHY_TCHLA_init: Initial concentration (ug/L)init_phyto_proportion: vector of the initial proportion of total chla that is each phytoplankton group intchla_components_varsrepresents. Must add up to 1.0
Observation uncertainty
obs_error_temperature_intercept: the component that is independent of the measurement magnitudeobs_error_temperature_slope: the component that linearly scales with the magnitude of the measurementobs_error_wq_intercept_phyto: Not usedobs_error_wq_slope_phyto: Not usedobs_error_wq_intercept: the component that is independent of the measurement magnitude. A vector of with the same length aswq_namesobs_error_wq_slope: the component that linearly scales with the magnitude of the measurement. A vector of with the same length aswq_names
Process uncertainty
temp_process_error: variance of process uncertaintyOXY_oxy_process_error: variance of process uncertaintyCAR_pH_process_error: variance of process uncertaintyCAR_dic_process_error: variance of process uncertaintyCAR_ch4_process_error: variance of process uncertaintySIL_rsi_process_error: variance of process uncertaintyNIT_amm_process_error: variance of process uncertaintyNIT_nit_process_error: variance of process uncertaintyPHS_frp_process_error: variance of process uncertaintyOGM_doc_process_error: variance of process uncertaintyOGM_poc_process_error: variance of process uncertaintyOGM_don_process_error: variance of process uncertaintyOGM_pon_process_error: variance of process uncertaintyOGM_dop_process_error: variance of process uncertaintyOGM_pop_process_error: variance of process uncertaintyNCS_ss1_process_error: variance of process uncertaintyPHS_frp_ads_process_error: variance of process uncertaintyPHY_TCHLA_process_error: variance of process uncertaintyPHY_process_error: variance of process uncertainty
Initial condition uncertainty
temp_init_error: variance of initial temperatureOXY_oxy_init_error: variance of initial oxygenCAR_pH_init_error: variance of initial pHCAR_dic_init_error: variance of initial DICCAR_ch4_init_error: variance of initial CH4SIL_rsi_init_error: variance of initial SilicaNIT_amm_init_error: variance of initial AmmoniumNIT_nit_init_error: variance of initial NitratePHS_frp_init_error: variance of initial PhosphorusOGM_doc_init_error: variance of initial DOCOGM_poc_init_error: variance of initial POCOGM_don_init_error: variance of initial DONOGM_pon_init_error: variance of initial PONOGM_dop_init_error: variance of initial DOPOGM_pop_init_error: variance of initial POPNCS_ss1_init_error: variance of initial SS1PHS_frp_ads_init_error: variance of initial Adsorped PhosphorusPHY_TCHLA_init_error: variance of initial Total Chl-aPHY_init_error: variance of initial Phytoplankton biomass
Management specific variables
simulate_SSS: include SSS (bottom-water oxygenation) in simulations with observed drivers (i.e., data assimilation simulations)TRUE: includeFALSE: don’t include
forecast_no_SSS: Include SSS in forecastTRUE: includeFALSE: don’t include
use_specified_sss: Use sss inflow and oxygen from file in forecast. IfFALSEthen provideforecast_SSS_flowandforecast_SSS_Oxy.forecast_SSS_flow: Flow rate of SSS in forecast (m3/day)forecast_SSS_Oxy: Oxygen concentration of SSS in forecast (mmol/m3)sss_fname: full path to the file that has the SSS Flow and oxygen datasss_inflow_factor: a scalar to multiply FLOW rate of SSSsss_depth: The depth (meters) of the SSS inflow/outflow
Plotting related options
focal_depths_manager: A vector of depths that are included in the plot with the % change of turnover.turnover_index_1: the top depth used in the calculation of whether the lake is mixedturnover_index_2: the bottom depth used in the calculation of whether the lake is mixed